Why did the Court of Appeal reverse the local division injunction in 10x Genomics vs Nanostring?
In the first ever public hearing of the Court of Appeal of the Unified Patent Court, the provisional injunction obtained by 10x Genomics against NanoString during first instance proceedings has been overturned. Previously the first instance court found that the patent in suit EP4108782B1 (EP’782) was valid with “a sufficient degree of certainty”, whereas the Court of Appeal ruled that that the patent was likely invalid due to a lack of inventive step.
The decision shows the UPC Court of Appeal employing some EPC concepts while not using others, e.g. the problem-solution approach of the EPO, which the lower court at least considered. However, it is nonetheless only a decision on the provisional injunction, and no final decision from the merits has yet been issued by the UPC at either instance.
Here we have focussed on the technical aspects of the technology and how the Court of Appeal came to such a different decision from the first instance.
The novelty/inventive step attack
During first instance proceedings, NanoString filed Goransson et al., 2008 (“D6”) to attack the novelty of EP’782, an argument dismissed by the Munich local division. During the appeal, NanoString additionally alleged that combining the teachings of D6 with Stougaard et al., 2007 (“B30”) rendered EP’782 obvious. This argument found favour with the Court of Appeal.
EP4108782B1
EP’782 describes a tool which can analyse multiple target biomolecules simultaneously in a cell or tissue. Detection of multiple targets concurrently (“multiplexing”) is valuable as operators can interrogate a single sample multiple times, thereby requiring less input material and increasing efficiency. For example, a hospital could obtain a single sample from a patient with an unknown infection and identify the infectious agent through a single test.
The ‘detection reagents’ of the patent are composed of a targeting ‘probe’ and a nucleic acid ‘label' (Figure 1A). The probe targets one specific biomolecule such as a DNA or RNA sequence. The label contains multiple sites for subsequent detection. One single probe can be linked to many labels to enable signal amplification. The labels are recognised by decoder probes, so-called ‘SeqTags’, which contain a detection means such as a fluorophore. Different sets of SeqTags are added and imaged sequentially to produce a unique fluorescent signature specific to a detection reagent (Figure 1B).
Figure 1
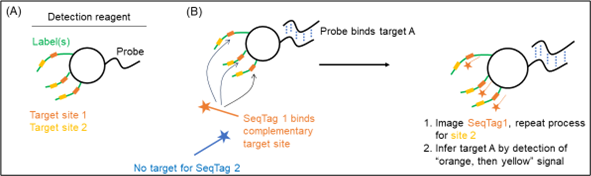
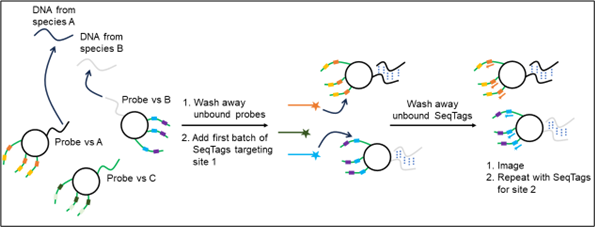
Bound detection reagents are retained and detected by the fluorescence of bound SeqTags, whereas unbound detection reagents are washed away. In Figure 2, species A is detected as first an orange signal then yellow, and species B is detected as blue signal then purple.
Figure 2
D6
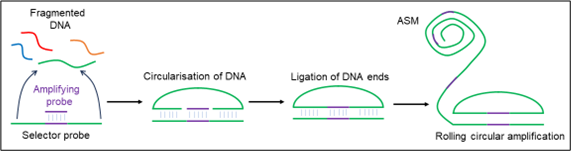
D6 describes a multiplexed method for detecting DNA sequences. A sample, such as fragmented genomic DNA, is mixed with a pool of ‘selector probes’ (Figure 3) which have 5’ and 3’ complementary regions for binding. These regions form a circular construct with complementary DNA. This is then ligated with an amplifying probe between the sample’s 5’ and 3’ ends. This circular construct is then amplified through rolling circle amplification to produce a long, concatemeric DNA product containing around 1200-2000 copies of the fragment. These products are termed “amplified single molecules” (ASMs).
Figure 3
The ASMs are immobilised onto a microscopy slide and a mixture of ‘sandwich probes’ and ‘tag probes’ are added (Figure 4). Each sandwich probe hybridises to an individual ASM and also contains two detection sites which bind tag probes. Tag probes bind to a specific sandwich probe at a specific detection site and contain a particular fluorophore.
Figure 4

Sets of sandwich and tag probes are added sequentially and imaged through fluorescence microscopy, with each set being dehybridised and washed off before the next is applied.
As with EP’782, each ASM can be identified through the specific fluorescence signal set. For example, a target may be defined by the detection of red signal at the first detection site on the first hybridisation, and then blue signal at the second detection site on the fourth hybridisation (Figure 5).
Figure 5

Conflict of interpretation
The Munich local division dismissed the relevance of D6. Claim 1 of EP’782 is directed towards detection “in a cell or tissue sample”. The ASMs are not cell or tissue samples. On the face of it, the technology seems quite different, but the claim is formulated more broadly in respect of the detection probes than the specific embodiment described in the patent.
In considering inventive step, the local division judged that the detection reagents remain bound to the biomolecule of interest throughout the detection procedure (feature (d) of claim 1). While this is not explicitly stated in the claims, they inferred this from “sufficient time to allow binding” in the incubating feature (c) and because reversal of this binding did not appear technically meaningful in EP’782. In D6, the detection probes are removed after each hybridisation step.
The Court of Appeal disagreed and took a more literal approach to the claims. They considered that claim 1 does not preclude that the decoder probes are removed after they have bound and that claim 1 does not require that (c) incubating and (d) detecting steps may not be carried out multiple times. They also noted that, in the description, incubation times range from 30 seconds to 48 hours and therefore it was “not impossible” for the detection reagents to be removed in between detection events.
The Court of Appeal therefore ruled that D6 disclosed all features except for “in a cell or tissue sample”. The discussion then became whether EP’782 was inventive over D6 for applying this method in situ. The Court of Appeal noted that D6 itself cited research on comparable multiplexed tools applied in situ, leading the skilled person to this subject matter directly.
B30 discloses a multiplexed method to detect RNA molecules in situ using similar technology. By combining these two documents, the Court of Appeal ruled that the skilled person would be capable of using the in vitro methods from D6 and applying it in situ to thereby render EP’782 obvious.
Conclusion
While both the local division and the Court of Appeal spent a great deal of time considering the technical details of the patented technology and that of the prior art, the lower court took a more technological approach to the interpretation of the claims. However, this may have resulted in reading limitations into the claims which were unjustified. Arguably, the local division dismissed D6 prematurely as being different from the patented technology, disregarding the broader terms in which the steps in claim 1 were defined.
By Stuart McKellar and George Lucas